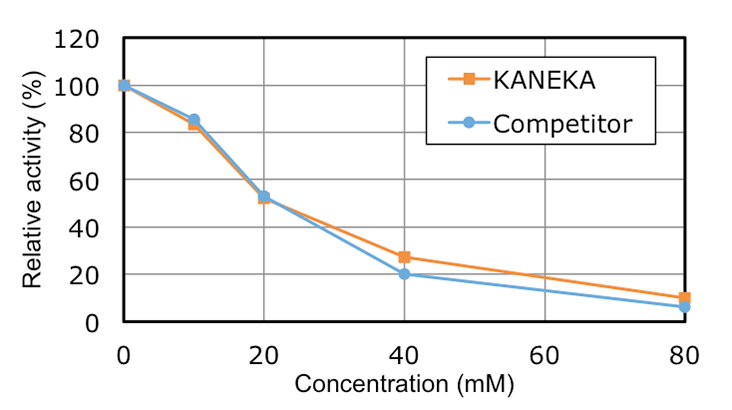
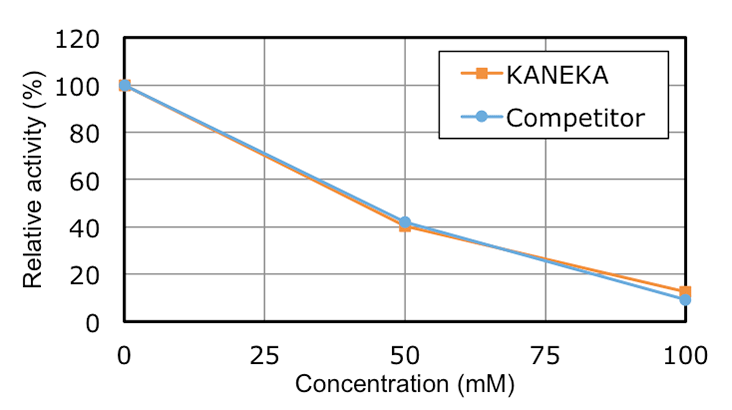
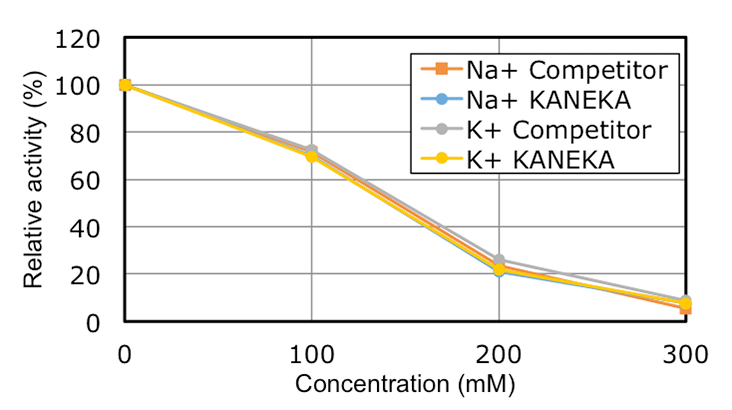
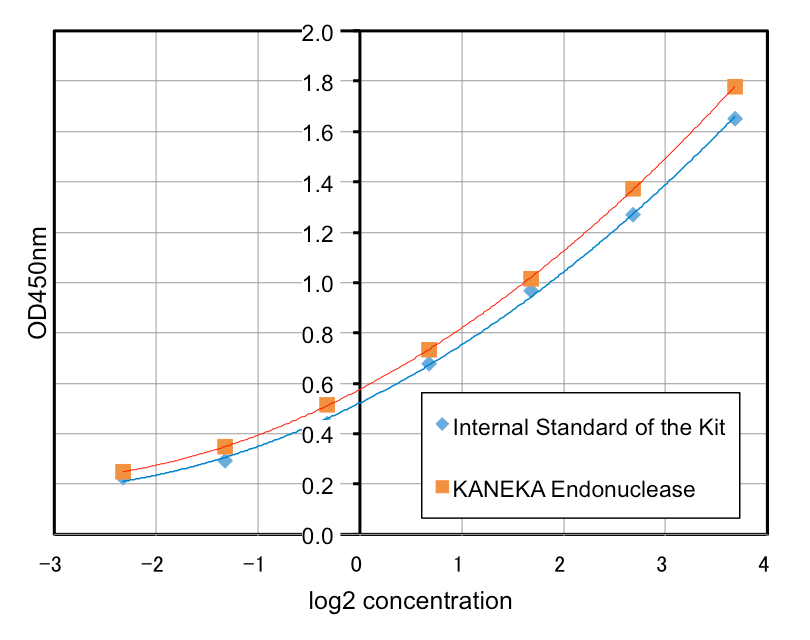
Characteristics
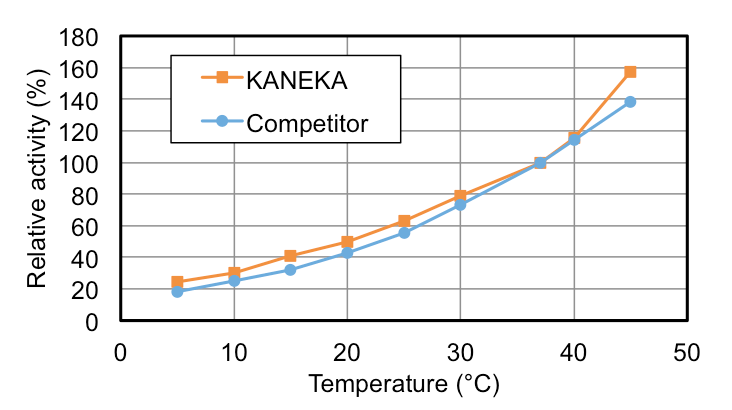
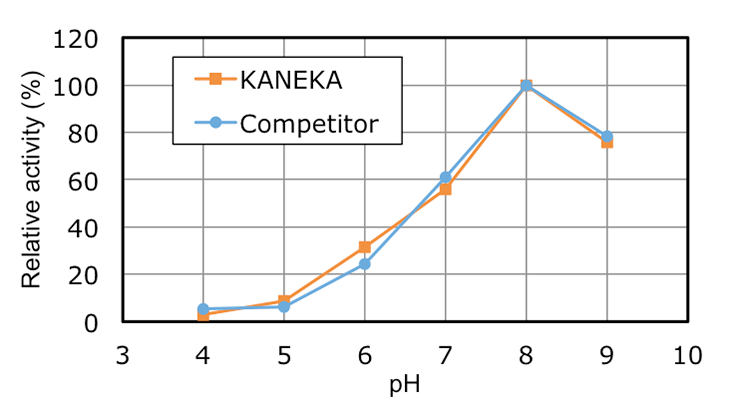
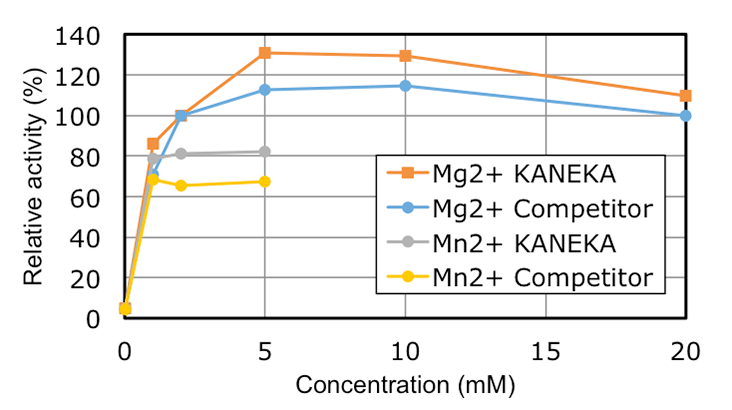
KANEKA Endonuclease is originating from the microorganism Serratia marcescens, and expressed in Pichia pastoris. A few amino acids substitution is introduced to avoid N-glycosylation, but it did not make any impacts on the original enzyme characteristics. KANEKA Endonuclease is a homodimer of 27 kDa subunit with a calculated pI of pH6.6, and Mg2+ is required for enzyme activity.




 About KANEKA Endonuclease
About KANEKA Endonuclease